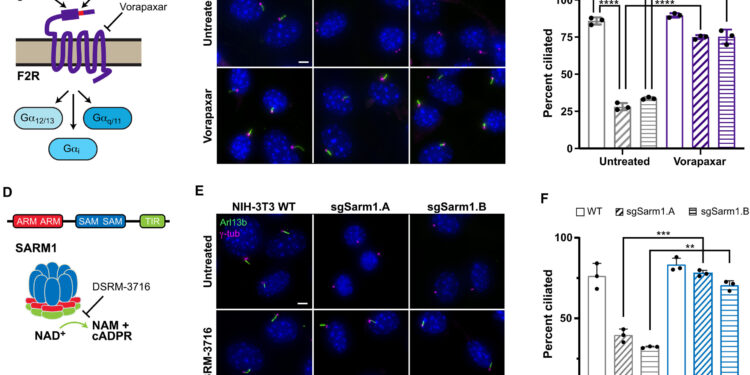
Overexpression of F2R or SARM1 induces cilia disassembly. (A) Illustration of F2R highlighting its activation, inhibition by vorapaxar, and signaling via G proteins. (B) Parental NIH-3T3 cells and cells transduced with F2r CRISPRa sgRNAs were serum starved for 24 hours, followed by immunofluorescence analysis of cilia (Arl13b) and centrioles (γ-tubulin (γ-tub)). Where indicated, cells were treated with 6 µM vorapaxar during serum starvation. (C) Quantification of ciliation from (B). Bars show means of N = 3 independent replicates (N > 75 cells each). (D) Schematic of SARM1, including the NADase TIR domain that cleaves NAD+ to produce nicotinamide (NAM) and cADPR. (E) Analysis of ciliation in NIH-3T3 cell lines as in (A). Where indicated, cells were treated with 30 µM DSRM-3716 during serum starvation. (F) Quantification of ciliation of (E). Bars show means of N = 3 independent replicates (N > 80 cells each). Credit: Scientific advances (2025). DOI: 10.1126/sciadv.aeb7238
In many cells in the human body, hair-like protrusions called cilia act like antennas, allowing cells to receive signals from their environment and other cells. As cells grow and divide, each cilium must first form on the cell body and then disassemble – or break down by breaking off or shortening – before the cell divides.
Many genes involved in eyelash formation have already been studied, and mutations in these genes are known to cause a host of pediatric disorders, or ciliopathies, that impact the formation of body systems like the skeleton, heart, and brain.
However, scientists know little about the genes that play a role in cilia disassembly, as well as how defects in the process impact the body.
In a new study, a Yale research team has identified a series of genes that constitute a pathway responsible for the disassembly of primary cilia (a type of single, immobile cilia found in certain cells) and, when defective, may be linked to a neurological disorder called focal cortical dysplasia.
The results are published in Scientific advances.
“Our goal was to use new genomic technologies to systematically address the question of cilia disassembly,” said David Breslow, associate professor of molecular, cellular and developmental biology in the Yale School of Arts and Sciences, and corresponding author of the study. “We discovered fundamental aspects of cell function that had not been well understood, as well as a potential new link to disease that could help us understand its causes and therapeutic strategies.”
Studying cilia disassembly could lead to better therapies for neurological diseases, as well as other conditions, including cancer, to which abnormal cilia breakdown may contribute.
Previously, Breslow said, members of his lab began using CRISPR (“clustered regularly interspaced short palindromic repeats”), a gene-editing technology, to identify genes needed by cells to build primary cilia. To do this, they neutralized (inhibited) the function of each gene in the genome, a technique that allowed them to link certain genes to diseases associated with defective cilia formation.
They then focused on a less studied process: eyelash removal.
“Here we turned this previous technology on its head,” said Breslow, who is also a member of the Wu Tsai Institute and the Yale Cancer Center. “Instead of asking which genes in the genome lead to defects in primary cilia when we inhibit their function, we now asked which genes impair primary cilia when we increase their function.”
To carry out this new type of genetic screening, the researchers used CRISPRa (CRISPR activation), a variant of CRISPR which allowed them to increase the activity of each gene in the genome. The scale of this experiment, Breslow said, was technically out of reach until recently.
Using this technique, the researchers were able to identify two genes, F2R and SARM1, that caused cilia loss when their activity increased (when overexpressed). Together, when functioning normally, these genes form a key pathway for cilia disassembly and help maintain normal ciliary function by regulating the balance of cilia assembly through disassembly.
Their second discovery, however, was more serendipitous.
Once researchers discovered this pathway, they began to wonder what impact its function might have on physiology. They found a possible answer in a research paper that identified the set of genes mutated in patients with focal cortical dysplasia, a neurodevelopmental disorder that causes seizures. To their surprise, the mutated genes included many of the same genes that made up the cilia disassembly pathway.
This overlap signals a potential key role that ciliary dysfunction may play in focal cortical dysplasia. This also highlights the possible physiological impacts of the cilia disassembly pathway identified by the researchers.
“Ultimately, by manipulating this pathway, we could develop treatments that would help restore cilia in diseases caused by excessive cilia disassembly, with cortical dysplasia potentially being an example,” Breslow said.
The team hopes to continue their research into cilia disassembly and the role that ciliary dysfunction plays in focal cortical dysplasia.
“We hope that new insights into cilia disassembly will help accelerate or catalyze future discoveries,” Breslow said. “There are some indications that cilia disassembly might be altered in neurological diseases, as we mentioned in this article, but perhaps also in other conditions, including cancer. This is something we are excited to explore further.”
More information:
Shane D. Elliott et al, A CRISPR activation screen reveals a mutated cilia disassembly pathway in focal cortical dysplasia, Scientific advances (2025). DOI: 10.1126/sciadv.aeb7238
Provided by Yale University
Quote: Pathway provides new insights into how cilia on cell surfaces break down before division (October 29, 2025) retrieved October 30, 2025 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.