Credit: Cell (2024). DOI: 10.1016/j.cell.2024.09.019
Researchers at Rutgers University in New Brunswick, along with international collaborators, have introduced a new method to identify the crucial set of gut microbes commonly found in humans and essential for health.
The researchers, whose study was published in Cell, said this discovery offers innovative opportunities for precision nutrition and personalized therapies aimed at managing chronic diseases associated with gut microbiome imbalances, including diabetes, inflammatory bowel disease and cancer.
The core microbiome refers to a collection of microbes found in the digestive tract that play an essential role in maintaining functions such as digestion, immune defense and mental health. When the core microbiome is reduced or lost, it can lead to a condition known as dysbiosis, an imbalance between beneficial and harmful microbes in the gut. Dysbiosis has been linked to many chronic diseases, including inflammatory bowel disease, metabolic disorders, neurological disorders, chronic kidney disease, and some cancers.
Many studies have shown that transferring beneficial fecal microbiota from a healthy to a diseased colon can alleviate these conditions, clearly indicating that a core microbiome is crucial for maintaining our health.
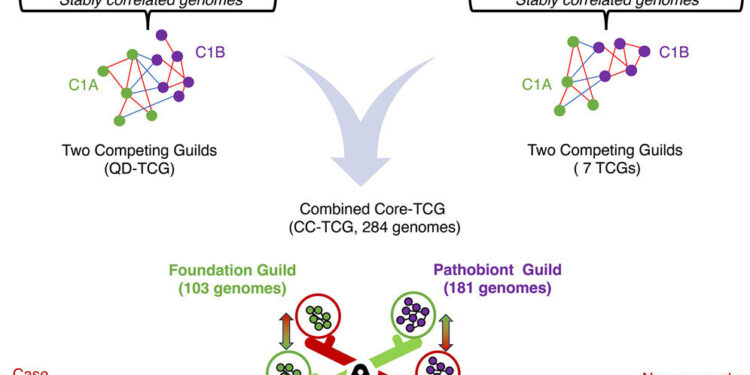
The essential structure of the core microbiome – two distinct groups of bacteria, called the foundation guild and the pathobiont guild – engage in dynamic and stable interactions that are crucial for maintaining human health. Using artificial intelligence models, the “two competing guilds” approach classifies cases from controls in diverse populations, unaffected by ethnicity, geography or disease types, and predicts personalized responses to immunotherapy in four different diseases.
The field has not yet reached consensus on what exactly constitutes the core microbiome or how to accurately identify these key microbial players.
Conventional methods of microbiome analysis often define the core microbiome using commonly shared taxonomic units, such as species or genus, within a human population. However, these taxa may have limited resolution. For example, within the same species, there can be strains that are both beneficial and harmful. The well-known intestinal bacterial species E. coli consists of mostly benign strains, but E. coli O157 can cause serious foodborne illnesses.
This new study overcomes these limitations by using high-quality genomes directly assembled from metagenomic sequencing datasets. Each genome is tagged with a universally unique identifier to track its ecological behavior. This genome-specific approach not only provides high resolution for analysis, avoiding mixing of signal with noise, but also includes the genomes of unclassifiable novel bacteria that are not limited by incomplete databases.
“Our research identifies bacteria in the gut that stay connected no matter what challenges the body faces, such as dietary changes or disease,” said Liping Zhao, Eveleigh-Fenton Professor of Applied Microbiology and professor in the Department of Biochemistry and Microbiology at the Rutgers School of Environmental and Biological Sciences. “By focusing on these resilient, interconnected bacteria, we have developed a new method to identify the microbes most essential to maintaining our health.”
This approach led to the identification of two distinct and opposing groups of essential gut bacteria: the beneficial founder guild and the necessary but potentially dangerous pathobiont guild.
The foundation guild is crucial for structuring and stabilizing the entire gut microbiome. These bacteria break down dietary fiber and produce short-chain fatty acids (SCFAs) such as butyrate, which are essential for gut health by supporting the intestinal barrier, reducing inflammation, and serving as an energy source for cells of the colon. SCFAs are also essential for suppressing harmful bacteria.
In contrast, the pathobiont guild, although necessary in small quantities for immune training and vigilance, can promote disease progression when it becomes ecologically dominant.
The seesaw balance between these two guilds is crucial. When the foundation guild dominates, gut health is preserved. However, when the scales tip in favor of the pathobiont guild, dysbiosis occurs, potentially leading to inflammation that can worsen various chronic diseases.
“Our model not only helps us identify these core bacterial guilds, but also shows how they can be nurtured to maintain their dominance,” Zhao said. “This opens up new possibilities for personalized nutrition and targeted therapies capable of restoring balance to the gut microbiome.”
By targeting fiber-degrading genes of the founding guild, personalized dietary recommendations can be formulated to support the ecological dominance of these key microbes.
The two competing guilds model provides both a new method and a new standard for identifying members of the core microbiome. By requiring that members of the core microbiome are not just shared within a population, but stably connected despite dramatic changes in their environment, the model sets a new benchmark for microbiome research, according to Zhao.
Zhao and his team plan to conduct a series of trials to further refine personalized therapies aimed at restoring and maintaining ecological dominance of the foundation guild in patients with severe dysbiosis. By applying the two competing guilds’ model to the clinical setting, they aim to translate their research into practical treatments that can significantly improve patient outcomes in conditions previously considered irreversible.
More information:
Guojun Wu et al, A central signature of the microbiome as an indicator of health, Cell (2024). DOI: 10.1016/j.cell.2024.09.019
Cell
Provided by Rutgers University
Quote: AI models help redefine the core microbiome for personalized therapies (October 16, 2024) retrieved October 16, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.