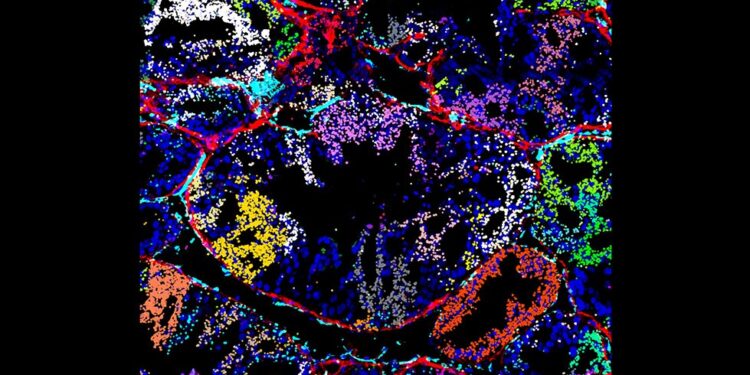
Visualization of in vivo detection of CRISPRmap barcodes on a xenograft tumor section, showing the distribution of the guide in the tumor microenvironment. Decoded CRISPR guide ID barcodes detected by CRISPRmap are displayed as false-colored dots based on their guide identity. Credit: Gaublomme Laboratory
The Gaublomme laboratory has developed a new clustered optical screening approach called CRISPRmap, which allows the optical properties of individual cells to be coupled to targeted genetic disruptions. Optical phenotypes are generally inaccessible for sequencing-based approaches based on cell lysis, but include crucial information such as cell morphology, subcellular localization of proteins, cell-cell interactions, extracellular matrix factors and tissue organization.
CRISPRmap enables spatially resolved interrogation of gene function in tissues, allowing researchers to map the cellular intrinsic and extrinsic effects of disruptions, which are not accessible via in vitro studies. This approach opens new avenues to understand genes involved in immune cell recruitment in tumors, metastasis, invasion, angiogenesis, etc.
The Gaublomme group shared its findings in a study recently published in Natural biotechnology.
Performing these studies in a pooled manner allows for high-throughput genetic studies by measuring the responses of many cells to different genetic perturbations in parallel. In pooled analyses, each cell expresses RNA transcripts that encode a barcode identifying the CRISPR disruption that occurred in that cell.
Notably, CRISPRmap enables optical barcode reading in challenging cell types and contexts previously elusive for other methods, including stem cells, neuron-derived cells, and in vivo cells in tissue contexts. After identifying which gene is disrupted in a given cell, scientists can learn more about how cells and their environment respond to that disruption.
“Our laboratory has optimized CRISPRmap to be compatible with optical readout assays, enabling simultaneous multiplexed profiling of proteins and mRNA species. Furthermore, CRISPRmap is independent of the type of genetic disruption, paving the way for the exploration of targeted mutations, gene interference and activation, epigenetic events. modification and editing of CRISPR RNA,” explained Professor Jellert Gaublomme, the corresponding author of the study.
In collaboration with the Ciccia laboratory at Columbia University Irving Medical Center, the authors applied CRISPRmap to study the functional consequences of 292 mutations in 27 genes essential for the DNA damage response, visualizing the recruitment of DNA damage proteins. DNA damage repair at sites of DNA damage.
They assessed these responses after ionizing radiation or DNA-damaging agents commonly used as chemotherapy drugs in breast cancer. Profiling the expression and subcellular localization of dozens of proteins and mRNA species in approximately one million cells has enabled nuanced interrogation of the diverse influences on the DNA damage response.
“This method allowed us to identify missense variants of uncertain clinical importance whose response resembles known pathogenic variants. As such, our approach can provide a framework for annotating human variants in a treatment-specific manner and help prioritize therapeutic strategies,” described Jiacheng Gu, the first author of the study.
Beyond studies of cancer cell lines grown in a dish, the researchers demonstrated their detection of CRISPRmap barcodes in cancer cells from the tumor microenvironment, a key goal of the NIH Director’s New Innovator Award that the lab received.
In collaboration with the Chan lab, they profiled tumor sections with CRISPRmap barcode detection and combined the readout with multiplexed antibody staining to visualize angiogenesis, extracellular matrix formation around tumor domains, and nuclear translocation of transcription factor in transplanted cells.
As CRISPRmap is applicable to various CRISPR modalities and cell types, the technique can be used for a wide range of research in biology and medicine. “We optimized our approach for broad accessibility because it does not rely on third-party sequencing reagents for barcode detection, the readout dyes can be customized to match the microscopes available to researchers, and the approach is profitable,” Gu said.
“We envision that this will allow individual laboratories to perform disruption studies on the cell type of interest, probing biological pathways on a scale similar to the number of genes known to play a role in the pathway.” added Gaublomme.
The Gaublomme laboratory plans to explore the impact of genetic disruptions on tissue architecture and the interaction between cells in complex microenvironments. Further studies could focus on patient-derived organoids to investigate gene function in tissue and disease-specific contexts.
“We envision CRISPRmap to elucidate optical phenotypes across a wide range of biological length scales, from molecular scales such as DNA damage breakage foci in a single nucleus to cellular reorganization in entire organs .
“The approach could be applied to studies ranging from fundamental biology to disease mechanisms and optimization of therapeutic approaches in development, neurodegenerative diseases and cancer,” Gaublomme concluded.
More information:
Jiacheng Gu et al, Mapping multimodal phenotypes to cell and tissue disruptions with CRISPRmap, Natural biotechnology (2024). DOI: 10.1038/s41587-024-02386-x
Provided by Columbia University
Quote: Scientists develop new method to study gene function in cells and tissues (October 14, 2024) retrieved October 14, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.