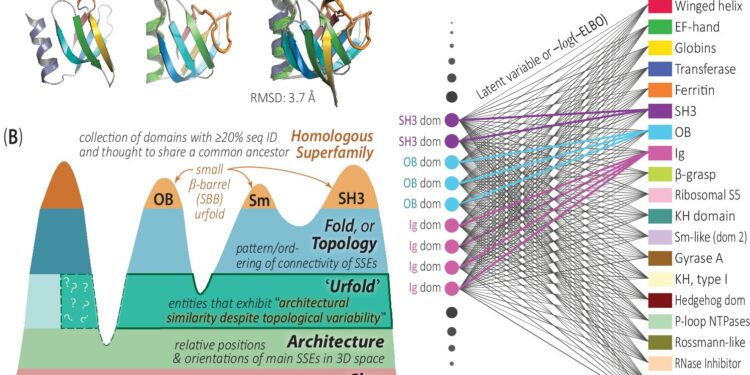
Presentation of the Urfold model and the DeepUrfold approach to identify domains likely to reflect the phenomenon of “architectural similarity despite topological variability”.
In an article recently published in Natural communications,a team proposes an AI-based approach to explore structural similarities and relationships in the universe of proteins. The team includes members from the University of Virginia, including Phil Bourne, dean of the School of Data Science, Cam Mura, senior scientist at the School, and Eli Draizen, a recent UVA alumnus.
Their study challenges conventional notions about protein structure relationships (i.e., patterns of similarities and differences) and, in doing so, identifies many weak relationships that elude traditional methods.
Specifically, the authors present a computational framework capable of detecting and quantifying such protein relationships on a large scale (across a myriad of proteins), in a novel, flexible, and nuanced way that combines deep learning-based approaches with a new conceptual model, known as Urfold. , which allows two proteins to exhibit architectural similarity despite different topologies or “folds”.
Bourne, Mura and Draizen collaborated on the project with Stella Veretnik. All authors are members of the Bourne & Mura Computational Biosciences Lab, part of UVA’s School of Data Science and Department of Biomedical Engineering.
This publication is the culmination of years of work by the Bourne Lab to develop this AI-driven framework, called DeepUrfold, to enable Urfold theory of structural relationships to be explored systematically and at scale.
Using DeepUrfold, the Bourne Lab team detected weak structural relationships in the protein universe between proteins that would otherwise have been considered unrelated, evolutionarily or otherwise.
By capturing and describing these distant relationships, DeepUrfold considers relationships between proteins in terms of “communities” and avoids the conventional approach of classifying proteins into separate, non-overlapping groups. Taken together, these new methodological approaches could push researchers to move beyond thinking about protein similarities in static and geometric terms and adopt a more integrated approach.
Bourne, founding dean of the School of Data Science, is known in the scientific community for his research, particularly in structural bioinformatics and computational biology in general. Earlier in his career, he co-led the development of the RCSB Protein Data Bank, a treasure trove of protein structure information that helped revolutionize the field and paved the way for contemporary AI advances like AlphaFold .
Mura, who holds positions in UVA’s School of Data Science and Department of Biomedical Engineering, has extensive experience in structural and computational biology, including biochemical and crystallographic studies of RNA-based systems and of the molecular biophysics of DNA. He views biological systems through the lens of molecular evolution and explores the intersection of these fields with data science.
Draizen earned a doctorate in biomedical engineering from UVA under Bourne’s mentorship and is currently a postdoctoral researcher in computational biology at the University of California, San Francisco. Veretnik has served as a senior research scientist at UVA focusing on computational biology and the structure, function and evolution of protein folds.
More information:
Eli J. Draizen et al, Deep generative models of protein structure discover distant relationships in continuous fold space, Natural communications (2024). DOI: 10.1038/s41467-024-52020-2
Provided by University of Virginia
Quote: An AI-based approach challenges traditional views on protein structure (October 10, 2024) retrieved October 10, 2024 from
This document is subject to copyright. Except for fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.