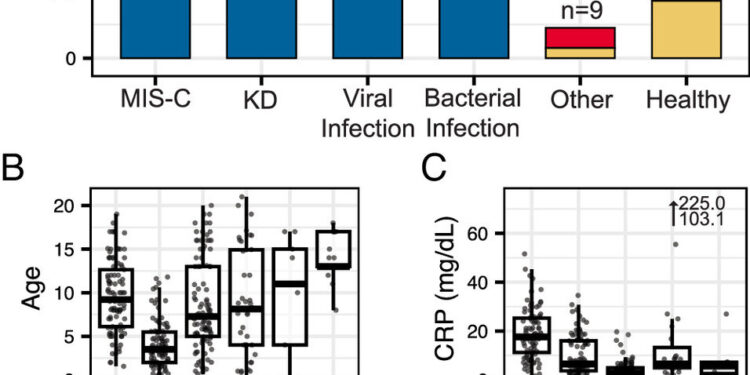
Sample overview. (A) Number of samples and distribution of originating hospital for each disease group. “Other” indicates other hospitalized pediatric controls. (B) Age distribution, (C) C-reactive protein (CRP) level, (D) Sex distribution, and (E) ICU status distribution for each sample group.
RNA serves many biological functions, from transmitting genetic information to making proteins. But RNA is also expelled from cells through cell death or active release, and can then end up in blood plasma.
A Cornell-led collaboration has developed machine learning models that use these cell-free molecular RNA residues to diagnose difficult-to-differentiate pediatric inflammatory diseases. The diagnostic tool can accurately determine whether a patient has Kawasaki disease (KD), multisystem inflammatory syndrome in children (MIS-C), a viral infection, or a bacterial infection, while simultaneously monitoring the health of the patient’s organs.
The article is published in the journal Proceedings of the National Academy of Sciences.
Inflammatory diseases pose a particular threat to children because symptoms, such as fever and rash, are generic and patients are often misdiagnosed. If not properly treated, MIS-C can cause swelling of the heart, lungs, brain, and other organs. Similarly, Kawasaki disease, the leading cause of acquired heart disease in children, can lead to heart aneurysms and heart attack. A cell-free RNA-based test would be the first molecular diagnostic tool that clinicians could use to detect these inflammatory diseases at a crucial early stage in children.
The Cornell team was led by Iwijn De Vlaminck, associate professor of biomedical engineering and co-senior author of the study. The lead author is Conor Loy, currently an Ignite Fellow for New Ventures.
These findings are the result of a previous collaboration that began four years ago and used next-generation sequencing to characterize severe cases of COVID-19 and MIS-C in children that increased during the pandemic.
Initially, De Vlaminck and Loy focused on the potential of cell-free DNA to study diseases, but they became increasingly interested in cell-free RNA, because of the wealth of information it provides. Although cell-free RNA has proven to be an effective biomarker for pregnancy and cancer, it is not as well studied as cell-free DNA.
“When you analyze RNA in plasma, you’re looking at RNA from dying cells, as well as RNA released from cells anywhere in the body,” Loy said. “That gives you a huge advantage. In inflammatory conditions, cell death is significant. In some cases, cells explode and their RNA is released into the plasma. By isolating that RNA and sequencing it, we can discover biomarkers of disease and trace the RNA back to the source to measure cell death.”
The collaborators studied 370 plasma samples from pediatric patients with various inflammatory diseases. The team converted the RNA into DNA, then performed DNA sequencing, which analyzed protein-coding regions of the genome. Loy spent a year experimenting with machine-learning algorithms to find disease signatures in the samples, creating a pipeline of different tools to make sense of the cell-free RNA.
Beyond developing an accurate model for diagnosis, the researchers also demonstrated that cell-free RNA sequencing can be used to quantify injury to specific tissues and organs, including the liver, heart, endothelium, nervous system and upper respiratory tract.
“I think a lot of the novelty and technical innovation lies in the data analysis,” De Vlaminck said. “We are able to quantify the amount of RNA coming from different organs, such as the liver or the epithelial cells of the vascular system. By quantifying the sources, we can also learn more about injury processes that are probably related to the immune system but that occur in vascularized tissues.”
At the same time, much is still unknown about cell-free RNA.
“We have shown that cell-free RNA is useful, but now we feel compelled to understand a little bit more about the biology of where the molecules come from and, perhaps more importantly, why they can be reproducibly isolated from plasma and why they are stable in plasma,” De Vlaminck said. “It’s because they are protected. We think we are starting to understand that as well.”
More information:
Conor J. Loy et al, Plasma cell-free RNA signatures of inflammatory syndromes in children, Proceedings of the National Academy of Sciences (2024). DOI: 10.1073/pnas.2403897121
Provided by Cornell University
Quote:Diagnostic tool identifies puzzling inflammatory diseases in children (2024, September 9) retrieved September 9, 2024 from
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without written permission. The content is provided for informational purposes only.