Application of splicing dependency models to cancer patient samples to prioritize splicing vulnerabilities with therapeutic potential. Credit: Nature Communications (2024). DOI: 10.1038/s41467-024-51380-z
Researchers from the Center for Genomic Regulation (CRG) have discovered hundreds of new genes potentially responsible for cancer. The results, published in the journal Nature Communicationssignificantly expands the list of possible therapeutic targets for monitoring and combating the disease.
Genetic mutations are the leading cause of cancer. They can change the shape and function of proteins, altering the normal functioning of a cell. According to COSMIC, the world’s most widely used cancer mutation database, 626 genes, when mutated, cause uncontrolled cell growth and survival. These are key therapeutic targets.
The study predicts that non-mutational mechanisms are just as prevalent. The researchers used an algorithm to find 813 genes that help cancer cells proliferate through alterations in an often-overlooked molecular mechanism called splicing. As with mutations, splicing can be targeted by drugs to control disease progression.
“By considering non-mutational mechanisms like splicing, we believe there could be twice as many potential genetic targets to control cancer. These are not classic oncogenes, but rather a whole new class of potential cancer drivers that can be targeted in isolation or in synergy with existing strategies. This is an incredibly exciting new frontier to explore,” says Miquel Anglada-Girotto, co-corresponding author of the study and a PhD student at CRG.
The study found that there was little overlap between genes involved in cancer development through splicing and genes involved in cancer mutation. Only about a tenth of the genes identified in the study (74, or 9.1%) are also present in the COSMIC database. The vast majority (508, or 62.5%) are potential cancer-causing genes that have been overlooked because they do not fit the traditional mutation-centric model of the disease.
“This tells us that splicing could be a largely independent mechanism driving cancer, complementary to well-known mutational pathways. It also explains why these potential targets have been historically overlooked, as cancer research has focused primarily on mutations,” Anglada-Girotto adds.
An algorithm called a “spotter”
Splicing is a mechanism that is often hijacked by cancer. When normal cells make proteins, they first copy the DNA of genes and create a first draft of instructions. Cells use splicing to remove unnecessary parts of the draft (introns) and glue together important pieces of information (exons).
Cancer cells include or exclude specific exons during splicing to create different versions of a protein from a single gene, some of which can promote cancer growth, survival, or drug resistance. This helps the cancer adapt to different environments or stresses, making it more aggressive and harder to treat.
Researchers have traditionally focused on specific splicing events or genes already suspected of being involved in cancer. The current study took a broader, “unbiased” approach, systematically analyzing splicing across the entire genome to identify new splicing events potentially responsible for cancer.
The researchers created an algorithm called spotter. The model looked at vast amounts of genetic data to spot the exons that cancer cells choose when splicing to promote growth. Spotter analyzed data from many different types of cancer cells to spot exons that are important for cell survival.
“Not only can Spotter identify potentially cancer-causing exons, which we can then link to genes, but it can also rank which exons are more important than others in a given cancer sample. We can use this to validate each exon experimentally so that the predictions made by the algorithm are confirmed,” Anglada-Girotto says.
Predicted protein structure of FNBP1, one of the genes potentially responsible for cancer. The study predicts that the inclusion of the exon (in yellow) helps cancer cells grow and spread. Credit: Miquel Anglada Girotto/Centro de Regulación Genómica
Testing predictions in the real world
While Spotter is a powerful tool for predicting genes that are potentially responsible for cancer through splicing, it is still just a prediction model. To see if its predictions hold up in real-world conditions, the researchers looked at a large dataset of nearly 7,000 samples from patients with 13 different types of cancer.
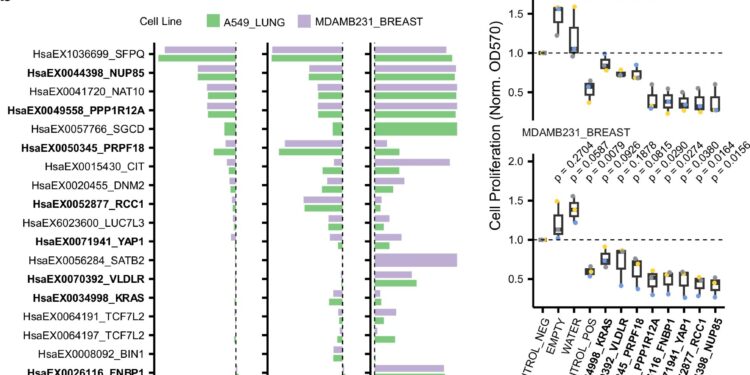
Splicing is known to play a larger role in aggressive, fast-growing cancers. The researchers used Spotter to test whether the algorithm could find the specific exons responsible. They used the algorithm to pre-select eight exons and designed synthetic drugs to target their splicing in cancer cell lines. As expected, the drugs were particularly effective at targeting fast-growing cancer cells.
“Spotter can help us go beyond the general understanding of the role of splicing in cancer to a much more detailed map of the specific parts of genes that are being hijacked by cancer cells. Essentially, it’s a way to find new, highly specific therapeutic targets,” says Dr. Luis Serrano, co-corresponding author of the research and director of the Center for Genomic Regulation.
Advancing “precision oncology”
The researchers also tested Spotter’s potential to predict cancer’s response to a drug. Changes in splicing can alter how a gene — and the protein it produces — responds to therapeutic molecules. The study explored how the splicing of certain exons can affect the sensitivity of cancer cells to these drugs.
The researchers combined the spotter’s predictions with data from large-scale experiments to identify exons linked to drug sensitivity. They used this data to create models that could predict how a cancer cell would respond to a particular drug. The researchers tested their model on data from 49 ovarian cancer patients and found that it could reliably distinguish between patients who were likely to be more resistant or more sensitive to chemotherapy.
“This could be part of a complementary strategy to understand the biology of a patient’s cancer and help oncologists determine the best risk-benefit ratio for cancer treatments and ultimately improve patient outcomes,” says Dr. Luis Serrano, co-corresponding author of the research and director of the Center for Genomic Regulation.
The researchers must overcome important limitations before their findings can translate into clinical applications. While the spotter can identify potentially cancer-causing exons, these predictions require extensive experimental validation to confirm their role in cancer. The study tested some predictions on cell lines, but the researchers will need to conduct broader validation on more cancer types and patient samples.
“Moving from computational predictions and cell line experiments to effective clinical treatments takes time and is fraught with challenges. However, because splicing has not been studied as extensively as mutations, there is still a vast uncharted territory to explore, ripe for new discoveries, some of which could change the way we think about and treat cancer,” concludes Dr. Serrano.
More information:
Miquel Anglada-Girotto et al, In silico screening of RNA isoforms to identify potential cancer driver exons with therapeutic applications, Nature Communications (2024). DOI: 10.1038/s41467-024-51380-z
Provided by the Center for Genomic Regulation
Quote: Hundreds of new cancer-causing genes predicted by algorithm (2024, September 3) retrieved September 3, 2024 from
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without written permission. The content is provided for informational purposes only.