Examples of inputs and outputs from the MADRC dataset. Credit: (2023). DOI: 10.7554/eLife.91398.1
Researchers have developed a suite of free tools to analyze large quantities of brain dissection photographs in brain banks around the world to improve understanding of neurodegenerative diseases.
The study, published today as a revised preprint in eLifeis described by the publishers as providing a valuable open source tool for researchers in the field of neuropathology and neuroimaging, supported by compelling evidence from experiments using both real and synthetic data.
Measuring the volume of different regions of the brain is an important way to understand aging and neurodegenerative diseases and is usually done either using magnetic resonance imaging (MRI) in people while they are alive or by studying sections of tissue brains donated to brain banks. after death.
Being able to relate the results of microscopic tissue analysis to large-scale macroscopic data from MRI scans is invaluable, but MRI scans are typically obtained on patients several years before their death, making it difficult to link this who is seen on the exam and what is observed. later seen under a microscope.
An alternative to this is to perform an MRI of the brain after the autopsy and before the tissue sections are taken for microscopic analysis. However, few biobank centers have the equipment or expertise to do this. Instead, quantitative measures such as cortical thickness and atrophy of specific regions are often estimated qualitatively by researchers examining brain slices.
“We decided to propose a solution to this problem by exploiting dissection photographs of brain slices routinely acquired before microscopic analysis,” says lead author Harshvardhan Gazula, a postdoctoral research associate at the Athinoula A. Martinos Center d Biomedical Imaging at Massachusetts General Hospital (MGH). ), Massachusetts, United States.
“These extensive collections of dissection photographs present an invaluable and currently underutilized information resource that promises to advance our understanding of diverse brain functions and disorders.”
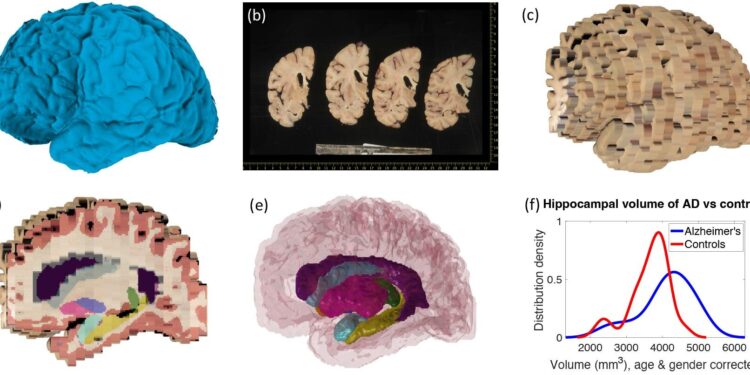
Harshvardhan and his colleagues have developed a suite of computational tools that will allow other researchers to use these dissection photographs to reconstruct a 3D image of the brain. The suite is available for free and includes three modules: the first corrects different perspectives and pixel sizes in the original images; the second constructs a 3D reconstruction of the brain from the photos, using a 3D analysis of the brain surface or a generic brain atlas as a reference point; and the third module then segments the brain reconstruction into different brain regions, such as the hippocampus, thalamus and cortex.
The structure and volumes of these regions can then be compared to other MRI scans and used alongside microscopic data to learn more about the changes that occur in neurodegenerative diseases.
To assess the accuracy of the tools, the team performed three validation steps. First, they used their tool to analyze dissection photographs of 21 post-mortem confirmed cases of Alzheimer’s disease and 12 age-matched control brains. This showed that the model captured well-known patterns of Alzheimer’s disease, such as shrinkage (atrophy) of the hippocampus and enlargement of the brain ventricle.
Next, they compared the accuracy of the new tool with the current benchmark analysis method, which uses a different technique for the segmentation step. This showed that the new tool outperformed the old version in several respects: for example, it was more effective at filling in missing information between brain slices.
Finally, the team assessed the robustness of the algorithms for reconstructing the brain on a large and variable set of simulated MRI data from 500 other participants. This showed that the method was reasonably robust even when there was more spacing between brain slice images, but it also showed that consistency in brain slice thickness is important for accuracy.
This study has some limitations, including that the method can only segment the entire cortex and is currently unable to subdivide it into regions, a task known as “cortical parcellation.”
Precise cortical parcellation would be of interest because the morphometry of individual regions, particularly measurements of their thickness, is more predictive of neuropathology. The authors say they are now looking to expand the toolset to enable cortical parcellation in the future.
“Harnessing the vast quantities of dissection photographs available in brain banks around the world to perform morphometry is a promising avenue for improving our understanding of various neurodegenerative diseases,” concludes lead author Juan Eugenio Iglesias, associate professor of radiology at the Athinoula A. Martinos Center for Biomedical Studies. Imaging at MGH.
“Our publicly available tools are easy to use by anyone with little or no training and provide a cost-effective and rapid link between morphometric phenotypes and neuropathological diagnosis. We hope that these tools will play a crucial role in the discovery of new imaging markers. to study neurodegenerative diseases.
More information:
Harshvardhan Gazula et al, Machine learning of dissection photographs and surface scanning for quantitative 3D neuropathology, eLife (2023). DOI: 10.7554/eLife.91398.1
eLife
Quote: New computer tools can reconstruct the brain in 3D from photos from the biobank (December 12, 2023) retrieved on December 12, 2023 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.