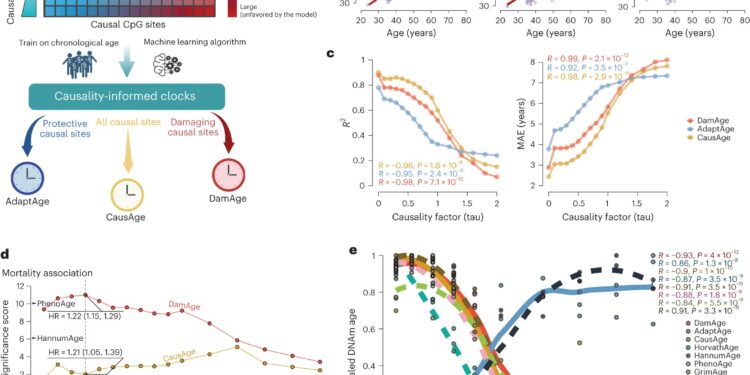
Construction and application of causally enriched epigenetic clocks. Credit: Natural aging (2024). DOI: 10.1038/s43587-023-00557-0
What makes us age? New “clocks” developed by researchers could help find answers. Investigators at Brigham and Women’s Hospital, a founding member of the Mass General Brigham Health System, are unveiling a new form of epigenetic clock: a machine learning model designed to predict biological age from DNA structure.
The new model distinguishes between genetic differences that slow and accelerate aging, predicts biological age and evaluates anti-aging interventions with increased precision. The results are published in Natural aging.
“Previous clocks considered the relationship between methylation patterns and traits that we know correlate with aging, but they don’t tell us which factors cause the body to age faster or slower. We created the first clock to make the distinction between cause and effect. said corresponding author Vadim Gladyshev, Ph.D., senior researcher in the Division of Genetics at BWH. “Our clocks distinguish between changes that accelerate and thwart aging in order to predict biological age and evaluate the effectiveness of anti-aging interventions.”
Aging researchers have long recognized the link between DNA methylation – alterations in our genetic structure that shape gene function – and its influence on the aging process. Notably, specific regions of our DNA, called CpG sites, are more strongly associated with aging. While lifestyle choices, such as smoking and diet, influence DNA methylation, so does our genetic inheritance, explaining why individuals with similar lifestyles can age at different rates.
Existing epigenetic clocks predict biological age (the actual age of our cells rather than chronological) using DNA methylation patterns. However, until now, no existing clock has distinguished between methylation differences responsible for biological aging and those simply correlated with the aging process.
Using a large genetic dataset, first author Kejun (Albert) Ying, a graduate student in the Gladyshev lab, performed epigenome-wide Mendelian randomization (EWMR), a technique used to randomize data and establish a causal link between DNA structure and observable traits. on 20,509 CpG sites causal to eight aging-related features.
The eight aging-related traits included lifespan, extreme longevity (defined as survival beyond the 90th percentile), health span (age at first incidence of serious age-related illness) , the frailty index (a measure of a person’s frailty based on the accumulation of health factors deficits over their lifetime), self-assessment of their health and three major measures related to aging incorporating family history, socioeconomic status, and other health factors.
With these features and their associated DNA sites in mind, Ying created three models, called CausAge, a general clock that predicts biological age based on causal DNA factors, and DamAge and AdaptAge, which include only damaging or protective changes. The investigators then analyzed blood samples from 7,036 individuals aged 18 to 93 from the “Generation Scotland cohort” and ultimately trained their model on data from 2,664 individuals in the cohort.
Using these data, the researchers developed a map identifying the human CpG sites responsible for biological aging. This map allows researchers to identify biomarkers responsible for aging and evaluate how different interventions promote longevity or accelerate aging.
The scientists tested the validity of their clocks on data collected from 4,651 people in the Framingham Heart Study and the Normative Aging Study. They found that DamAge was correlated with adverse outcomes including mortality, and AdaptAge was correlated with longevity, suggesting that age-related damage contributes to the risk of death while protective changes in methylation DNA can contribute to a longer lifespan.
Next, they tested the clocks’ ability to gauge biological age by reprogramming stem cells (transforming specialized cells, like skin cells, into a younger, less defined state where they can develop into different types cells in the body). When applying the clocks to newly transformed cells, DamAge decreased, indicating a reduction in age-related damage during reprogramming, while AdaptAge showed no particular pattern.
Finally, the team tested the performance of their clocks on biological samples from patients with various chronic diseases, including cancer and hypertension, as well as samples damaged by lifestyle choices like smoking. DamAge consistently increased in conditions associated with age-related damage, while AdaptAge decreased, effectively capturing protective adaptations.
“Aging is a complex process and we still don’t know which interventions against this phenomenon actually work,” Gladyshev said. “Our results present a step forward in aging research, allowing us to more precisely quantify biological age and assess the ability of new anti-aging interventions to increase longevity.”
More information:
Kejun Ying et al, Causally enriched epigenetic age dissociates damage and adaptation, Natural aging (2024). DOI: 10.1038/s43587-023-00557-0
Provided by Brigham and Women’s Hospital
Quote: New epigenetic clocks reinvent how we measure age (February 14, 2024) retrieved February 14, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.