Graphical summary. Credit: Angewandte Chemie International Edition (2023). DOI: 10.1002/anie.202313944
By attaching molecules together, scientists at the University of Montreal believe they have discovered how the molecular systems at the origin of life evolved to create complex self-regulatory functions.
Published in Modified chemistryTheir findings promise to provide chemists and nanotechnologists with a simple strategy for creating the next generation of dynamic nanosystems.
Life on Earth is supported by millions of different tiny nanostructures or nanomachines that have evolved over millions of years, explained Alexis Vallée-Bélisle, professor at UdeM and principal investigator of the study.
These structures, often less than 10,000 times the diameter of a human hair, are generally composed of proteins or nucleic acids. While some are made from a single component or part (often linear polymers that fold into a specific structure), most are made from multiple components that assemble spontaneously into large dynamic assemblages.
Respond to stimuli
“These molecular assemblies are very dynamic and activate or deactivate precisely in response to various stimuli such as a variation in temperature, oxygen or nutrients,” explained Vallée-Bélisle.
“Similar to cars that require sequential ignition, brake release, gear shifting, and throttle input to move forward, molecular systems require the sequential activation or deactivation of various nanomachines to perform specific tasks ranging from moving to breathing to thinking.”
The researchers raised a fundamental question: How were dynamic molecular assemblies created, programmed, and fine-tuned to support life?
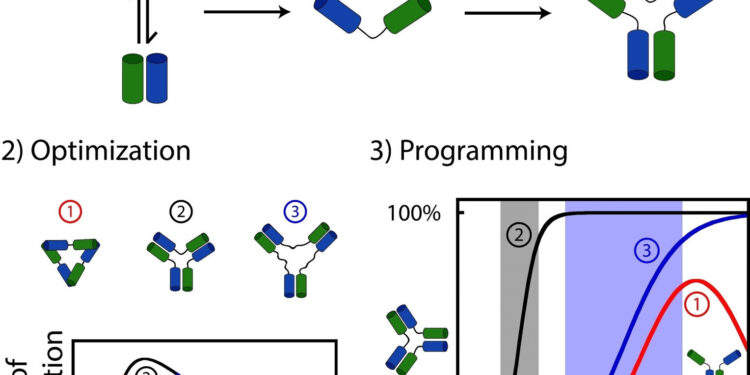
What they discovered was that many biological assemblies were likely formed by randomly attaching interacting molecules (e.g., proteins or nucleic acids such as DNA or RNA) with agents connection acting as a “connector” between each part.
“As these biomolecular assemblies play a crucial role in allowing living organisms to respond to their environment, we hypothesized that the nature of the connectivity between attached components could also contribute to the evolution of their dynamic responses,” he said. declared Vallée-Bélisle, holder of the Canada Research Chair in bioengineering and bio-nanotechnology.
Explore the impact of connectivity
To explore this question, Dominic Lauzon, a doctoral student at the time of the study, decided to synthesize and attach together dozens of interacting DNA molecules to explore the impact of connectivity on assembly dynamics.
“The programmable, easy-to-use chemistry of nucleic acids such as DNA makes them a practical molecule for studying fundamental questions related to the evolution of biomolecules,” said Lauzon, the study’s first author. “In addition, nucleic acids are also believed to be the molecule that gave rise to life on Earth.”
Lauzon and Vallée-Bélisle discovered that a simple variation in the length of the “linker” between interacting molecules leads to significant variations in their assembly dynamics. For example, some assemblages exhibited high sensitivity to stimulus variations, while others lacked such sensitivity, or even required much greater stimulus changes to promote assembly.
More surprisingly, some binding agents have even created new complex regulatory functions such as self-inhibition properties, where the addition of a stimulus would promote both its assembly and disassembly. All of these different reactive behaviors are also often observed in natural “living” nanomachines.
Using experiments and mathematical equations, the researchers were also able to explain why such a simple variation in linker length was so effective in changing the dynamics of molecular assembly.
“The linkers creating the most stable assemblies were those that also created the most sensitive activation mechanisms, while the linkers creating the least stable assemblies created the least sensitive activation mechanisms, even to the point of introducing the “self-inhibition,” Lauzon explained.
Detection is crucial
The ability to accurately detect molecular signals is crucial for biological assemblies but also for the development of nanotechnologies which depend on the detection and integration of molecular information.
The researchers therefore believe that their discovery could also provide the fundamental framework for creating more programmable nanomachines or nanosystems with optimally regulated activities, for example by simply attaching interacting molecules with different linkers. Such molecular assemblies already find applications in biosensing or drug delivery.
In addition to providing a simple design strategy for creating the next generation of self-regulated nanosystems, the scientists’ findings also shed light on how natural biomolecular assemblies might have acquired their optimal dynamics.
“A well-known molecular evolution strategy of living organisms is genetic fusion, where DNA encoding two interacting protein domains is fused randomly,” Vallée-Bélisle said.
“Our results also provide the fundamental understanding required to understand how simple variation in linker length between fused proteins may have effectively created biological assemblies exhibiting a variety of dynamics, some better suited than others to provide advantage to organisms. living.”
More information:
Dominic Lauzon et al, Principles of design and thermodynamics for programming the cooperativity of molecular assemblies, Angewandte Chemie International Edition (2023). DOI: 10.1002/anie.202313944
Provided by the University of Montreal
Quote: How the molecular systems at the origin of life could have evolved: the rise of nanomachines (February 7, 2024) retrieved on February 7, 2024 from
This document is subject to copyright. Apart from fair use for private study or research purposes, no part may be reproduced without written permission. The content is provided for information only.